In this post I’ll pretend that I am teaching a data science course on collecting, cleaning, storing, and updating data. Rather than pretending my students are computer scientists or software developers, I’ll pretend that they are business analysts or college grads going on to become business analysts. However, I’ll assume they know some programming (python), how to navigate their mac via the terminal, and they’ve read this post on installing mongodb as well as pymongo.
The chances are good that they will be spending much more time querying databases than building them. That doesn’t mean learning the process is a waste of time. At some point they will want to collect data that is not already collected and having an option other than a spreadsheet to store, update, and query that data will be a huge advantage. They can think of the data as information about customers or users, metadata about tweets, mobile phone models, or newspaper articles, but what we’ll actually demonstrate with is infobox data for different species of spiders.
At a high level here is what we are going to do.
- Grab a csv file from DBpedia detailing data on spiders and read that data into a python dictionary (we’ll worry about cleaning the data in a later post)
- Import the information into MongoDB
- Update our database with a new data field and query the database to see what we got
My recommended approach on how to do this and learn what’s happening is to do it in steps in an ipython console. I purposely kept the blocks of code out side functions to make this tutorial more fluid.
Get It, Clean It, & Prepare a List of Dictionaries
First things first, go to DBpedia and download the Species – Arachnid csv file and save as ‘spiders.csv’. We’ll want to get the information on each spider into a python dictionary and then we will append each one of these dictionaries to a list. This will allow us to easily import a list of dictionaries into mongo. To keep things simple we are only going to take a few fields — rdf-schema#label, synonym, rdf-schema#comment, family_lable, and phylum_label. Here is an example of what that process might look like…
import csv
DATAFILE = 'spiders.csv'
FIELDS = ['rdf-schema#label', 'synonym', 'phylum_label', 'family_label', 'rdf-schema#comment']
data = []
with open(DATAFILE, "r") as f:
reader = csv.DictReader(f)
# Skip first 3 rows bc they are metadata
for i in range(3):
l = reader.next()
# Iterate through each spider...
for line in reader:
temp = {}
# Iterate through each field of target spider
for field, val in line.iteritems():
if field not in FIELDS:
continue
# Only add required fields
if field in FIELDS:
temp[field] = val
# Append spider to our data list
data.append(temp)
Great. Now we have a list of 3,967 spider dictionaries (len(data)). To keep this simple (again) let’s write our list of dictionaries to a .json file. If you were wondering before why we used DictReader, it’s because mongodb’s foundation is the json object — which is very similar to a python dictionary. By using DictReader, we’ve made the next step extremely easy as you’ll see.
import json
with open('spiders.json', 'w') as outfile:
json.dump(data, outfile)
Into MongoDB
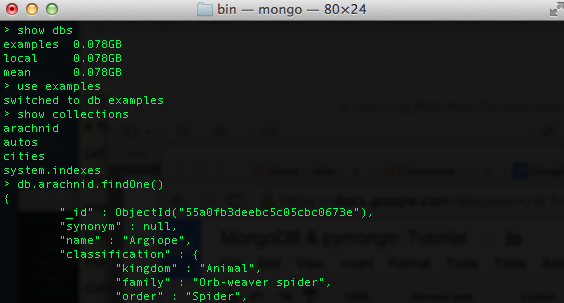
Fire up a mongod instance. If you forgot what that means, check out this post. We are going to take our spiders.json file… which is essentially a list of dictionaries… and use the mongo insert method to create 3,967 new entries in a collection called arachnid in a database called examples. As a quick reminder, in a mongo shell you can see your databases, collections, and look at a sample record using these commands…
Even if you do not yet have an examples database or an arachnid collection.. our python script will create one. So let’s have a look…
from pymongo import MongoClient
client = MongoClient("mongodb://localhost:27017")
db = client.examples
with open('spiders.json') as f:
data = json.loads(f.read())
db.arachnid.insert(data)
And voilà, we now have a database from which we can modify and run queries on. Let’s do that now.
Update and Search
So far so good. We’ve got some data and processed it into mongodb. Now the result of typing this in ipython…
db.arachnid.find_one()
Should look something like this…
{'family_label': 'Eriophyidae',
'phylum_label': '{Chelicerata|Arthropod}',
'rdf-schema#comment': 'Abacarus is a genus of acari including the following species: Abacarus acutatus Sukhareva 1985 Abacarus doctus Navia et al. 2011 Abacarus hystrix (Nalepa 1896) Abacarus lolii Skoracka 2009 Abacarus sacchari Channabasavanna 1966',
'rdf-schema#label': 'Abacarus',
'synonym': 'NULL'}
As a business analyst working in a dynamic environment with dynamic data we’ll need to assume that we’ll have to make changes to our records at some point in the future. As an example, let’s assume we want to add the spider’s genus to each record (this might be similar to adding a new type of address for customers (mailing address, email address, new kind of address we don’t yet know about???)). In order to do this we create a new list of dictionaries with our new fields of interest. This will be similar to above… however we only need to grab the genus value along with the label field (we need the label field so we can match it to the existing record in the database).
NEW_FIELDS = ['rdf-schema#label', 'genus_label']
data = []
with open(DATAFILE, "r") as f:
reader = csv.DictReader(f)
# Skip first 3 rows bc they are metadata
for i in range(3):
l = reader.next()
# Iterate through each spider...
for line in reader:
temp = {}
# Iterate through each field of target spider
for field, val in line.iteritems():
if field not in NEW_FIELDS:
continue
if field in NEW_FIELDS:
temp[field] = val
data.append(temp)
Next, we will loop through this list… match the label to an existing record in our database… and add the genus key and value to that record.
for el in data:
# Loop through the spiders in the new fields data list, match the label with existing spiders in database
spider = db.arachnid.find_one({'rdf-schema#label': el['rdf-schema#label']})
# Then add new genus field to that spiders data
spider['genus_label'] = el['genus_label']
db.arachnid.save(spider)
Good stuff. To be sure everything worked, use the same find_one method as we did above. Now, we should see a record with the genus key.
{u'_id': ObjectId('55a3099feebc5c09f32edf31'),
u'family_label': u'Eriophyidae',
u'genus_label': u'NULL',
u'phylum_label': u'{Chelicerata|Arthropod}',
u'rdf-schema#comment': u'Abacarus is a genus of acari including the following species: Abacarus acutatus Sukhareva 1985 Abacarus doctus Navia et al. 2011 Abacarus hystrix (Nalepa 1896) Abacarus lolii Skoracka 2009 Abacarus sacchari Channabasavanna 1966',
u'rdf-schema#label': u'Abacarus',
u'synonym': u'NULL'}
Summary
The above is one method for collecting, storing, and updating records. If students feel good with python, mongo’s python driver makes this process enjoyable. So what’s next? As we search through our database with mongo’s find and aggregate methods, we would notice that the data is a bit sloppy (synonyms are stored and separated like this { val | val } which makes iterating over them difficult, many NULL values, key names could be cleaner (use ‘name’ instead of ‘rdf-schema#label’), etc.). Making decisions on how to clean this data and programmatically implementing these solutions is a good next step for exploring MongoDB and pymongo. As an alternative option for a next step, we might look at the DBpedia ontology, selecting another dataset, and repeat the above procedure with that other data.